Tomato Leaf Disease Classification Using Custom CNN
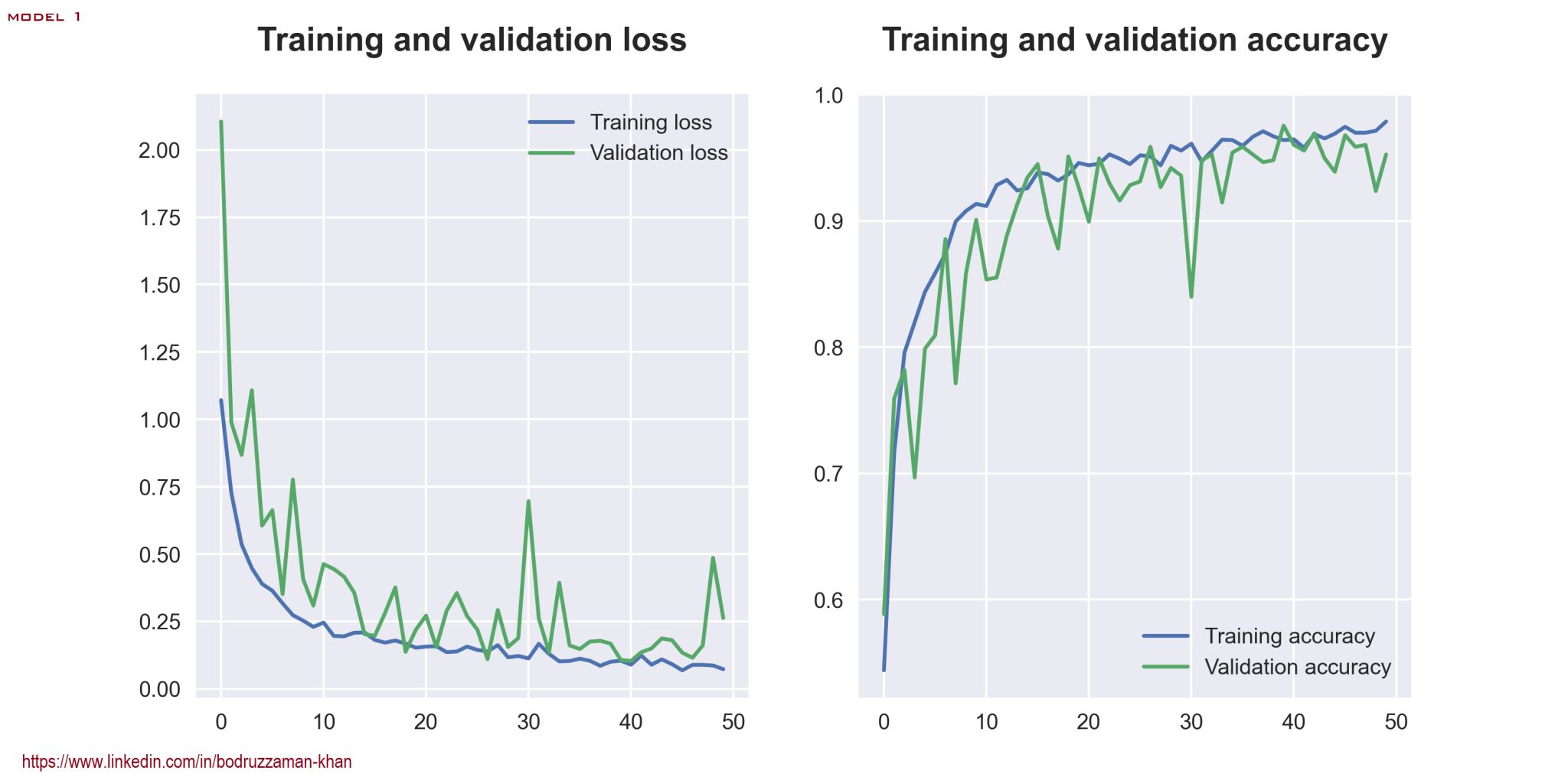
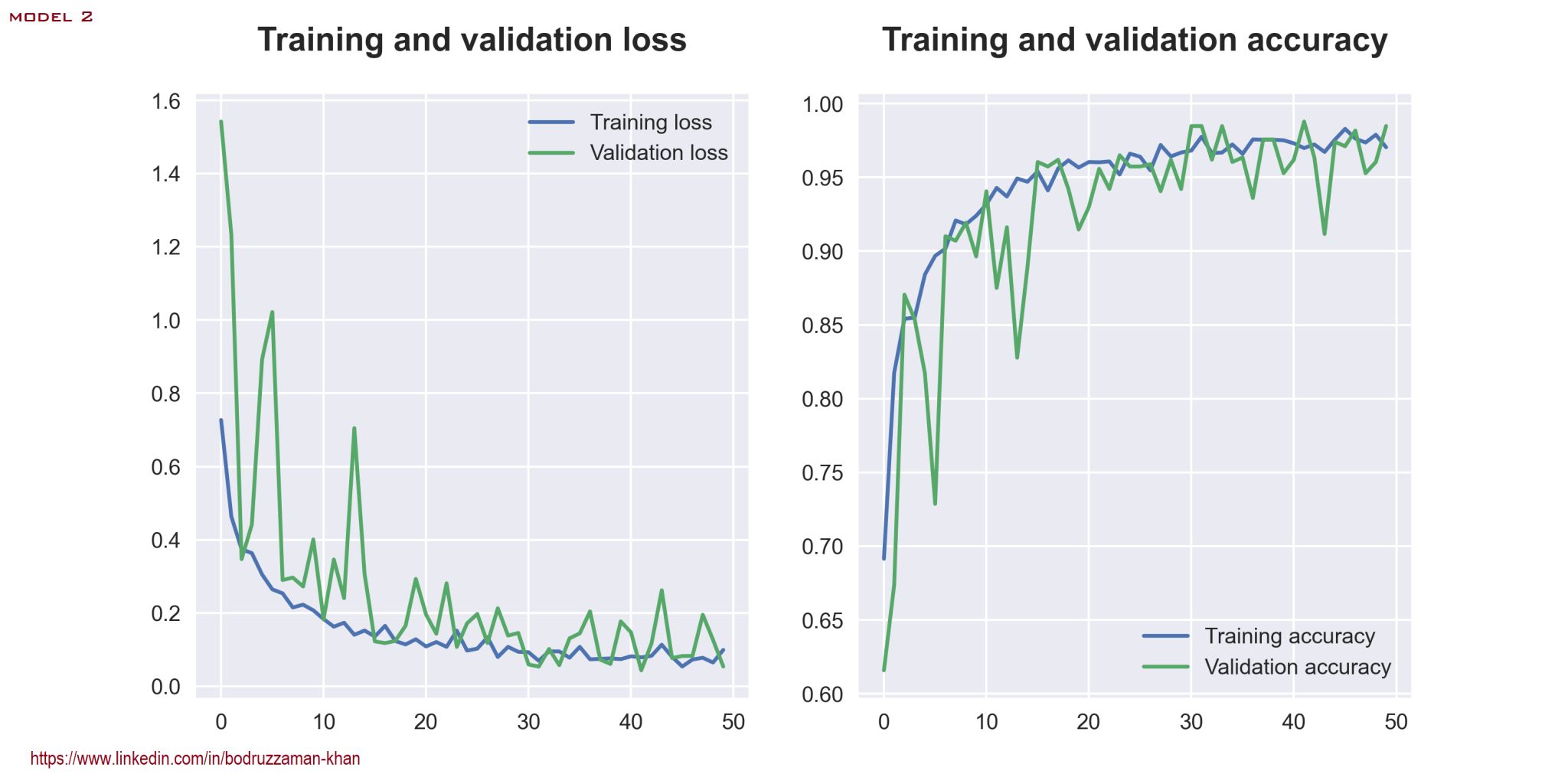
BackI have done a comparative analysis of 2 different CNN models for tomato leaf disease classification. Though there may be some issues regarding generalization, my models perform very well on test data. The first model has a loss of 0.2035 and an accuracy of 0.9509, while the second has a loss of 0.0562 and an accuracy of 0.9851 on test data. Some further tweaking might lead to a better result.
Model Architectures
Sample: 6627 units
1st Model Architecture
- A pre-processing layer that resizes and rescales the input images
- An augmentation layer that applies random transformations to the input images
- A 2D convolutional layer with 32 filters and a 3x3 kernel with ReLU activation function
- A 2D max pooling layer that reduces the dimensions by taking the max value of each 2x2 block
- A second 2D convolutional layer with 64 filters and a 3x3 kernel with ReLU activation function
- A second 2D max pooling layer
- A flatten layer that converts the output into a 1D vector
- A dense layer with 64 units and ReLU activation function
- A last dense layer with 4 units and a softmax activation function for class prediction
Loss and Accuracy: Model 1
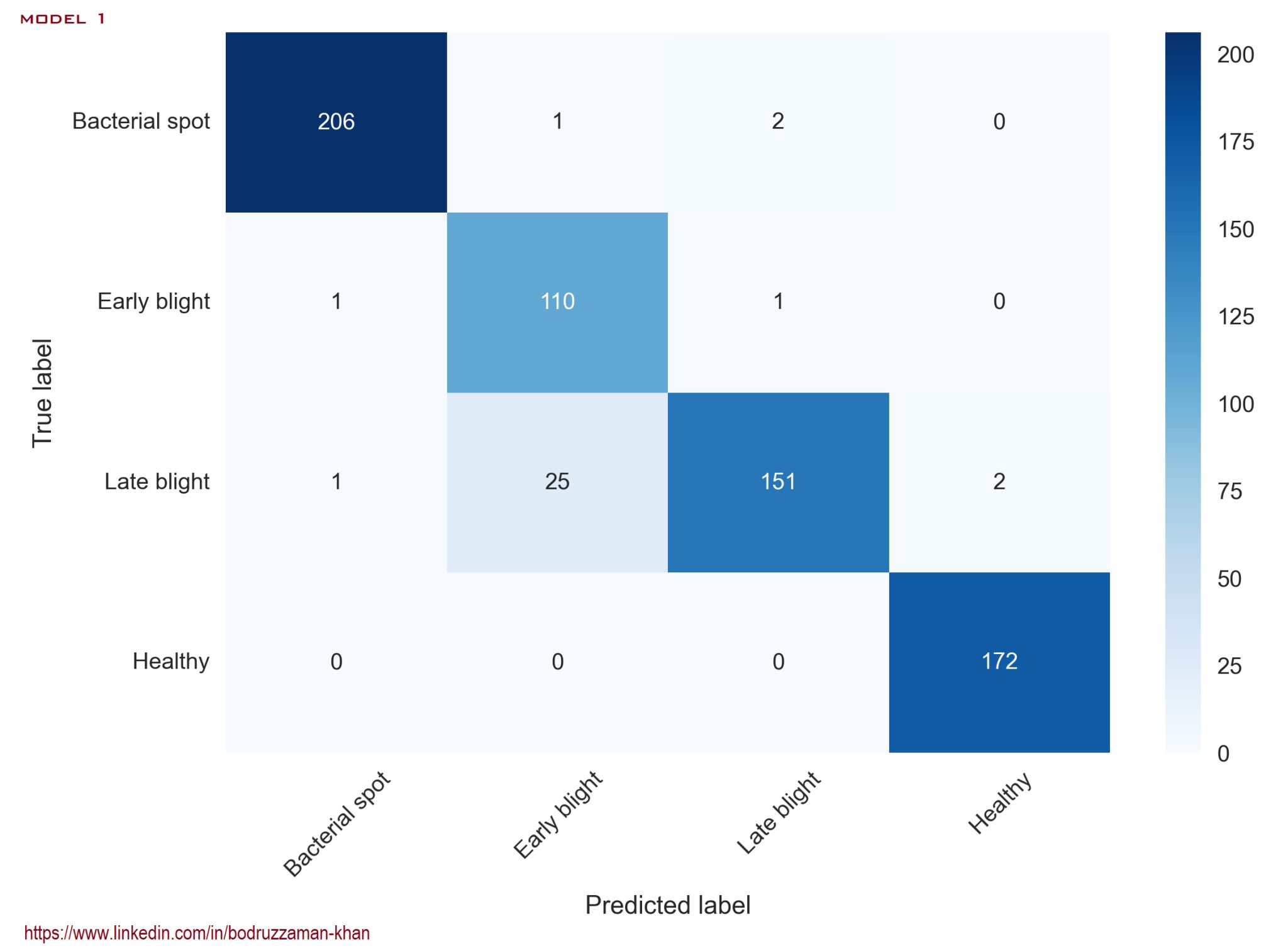
Confusion Matrix: Model 1
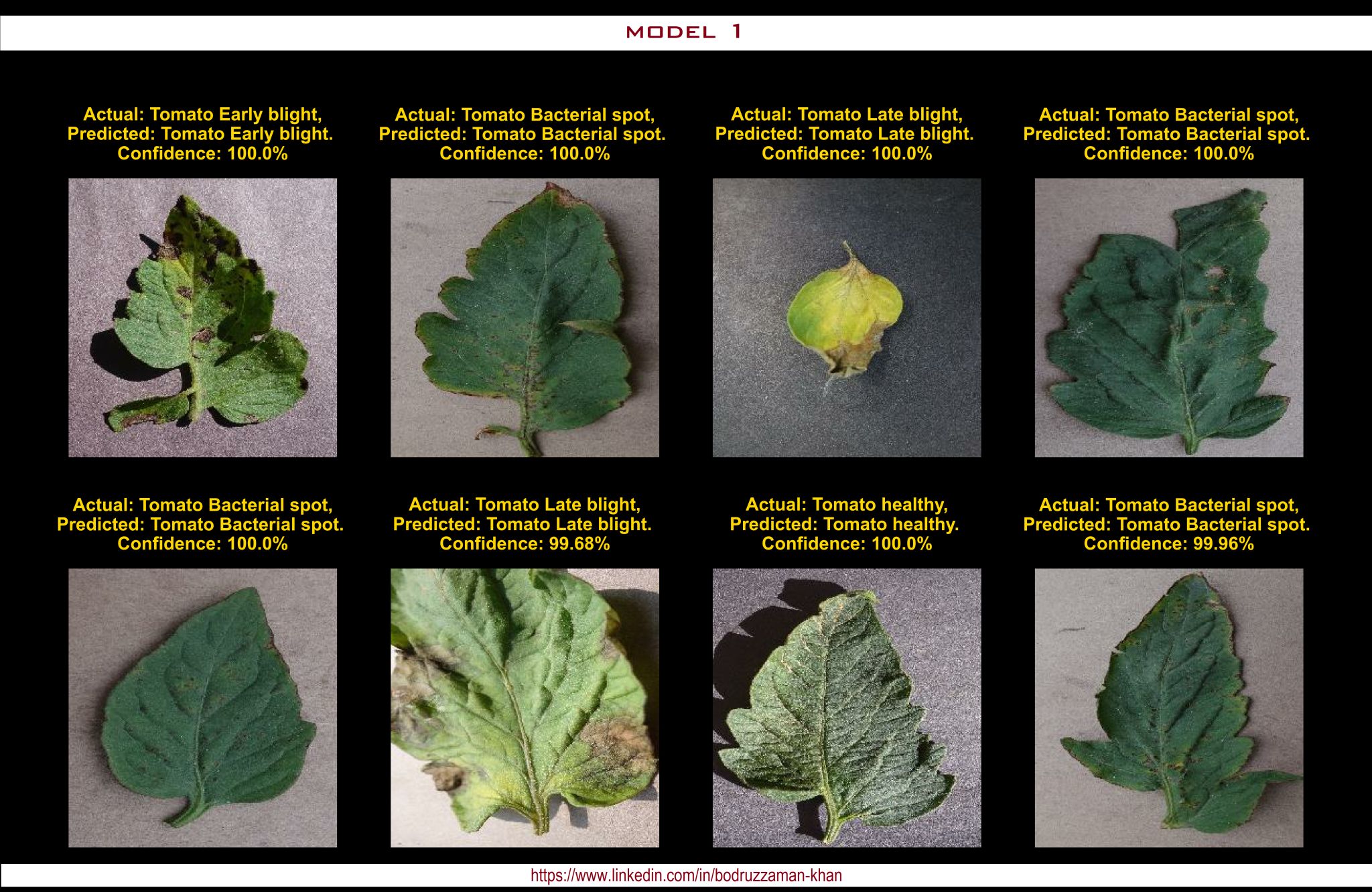
Classification Result: Model 1
2nd Model Architecture
Added layers:
- An additional convolutional layer
- An additional pooling layer
- Additional dense layers
Loss and Accuracy: Model 2
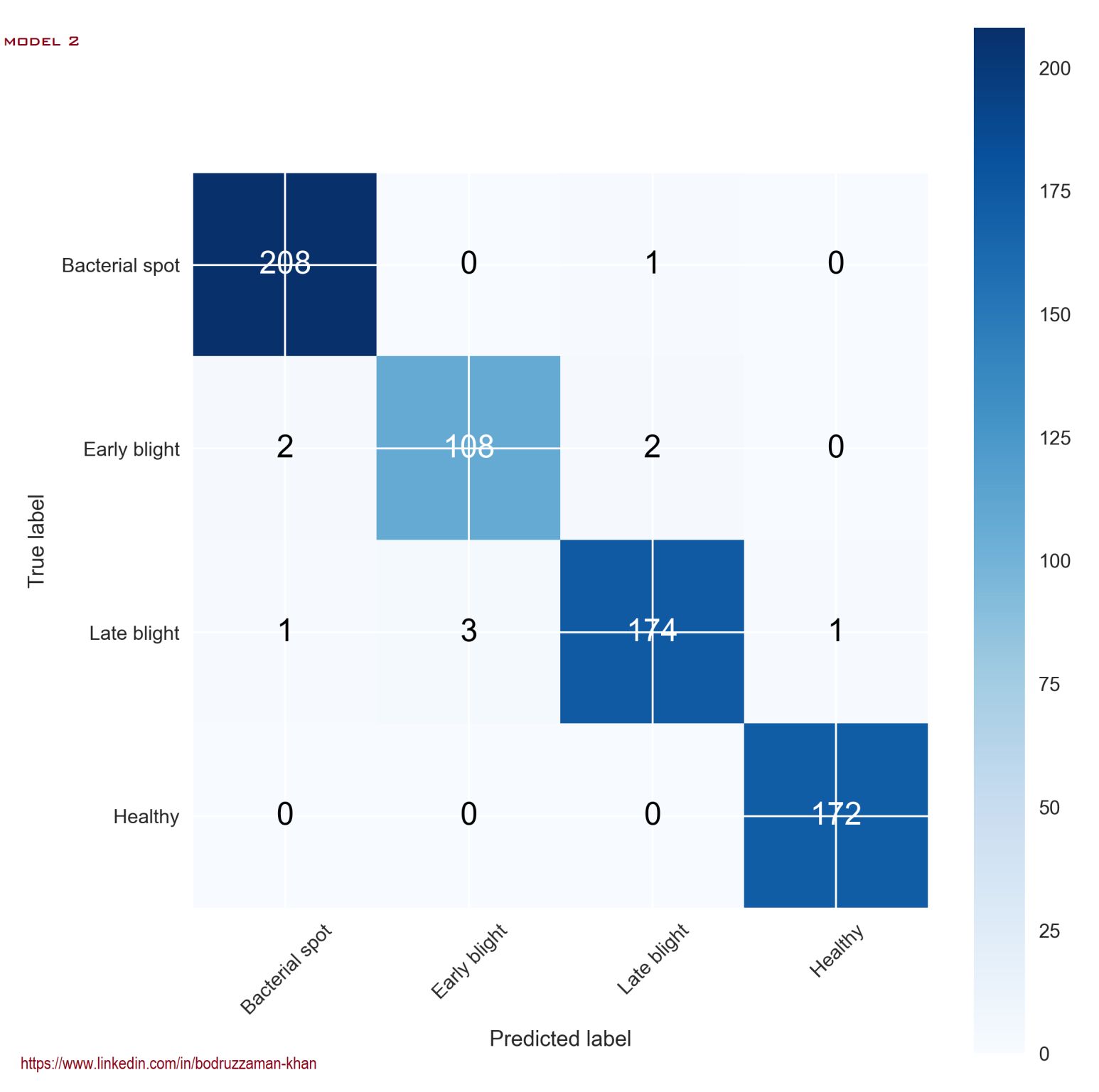
Confusion Matrix: Model 2
Classification Result: Model 2
